PEtab – a data format for specifying parameter estimation problems in systems biology

PEtab is a data format for specifying parameter estimation problems in systems biology. This repository contains the PEtab specifications and additional documentation.
About PEtab
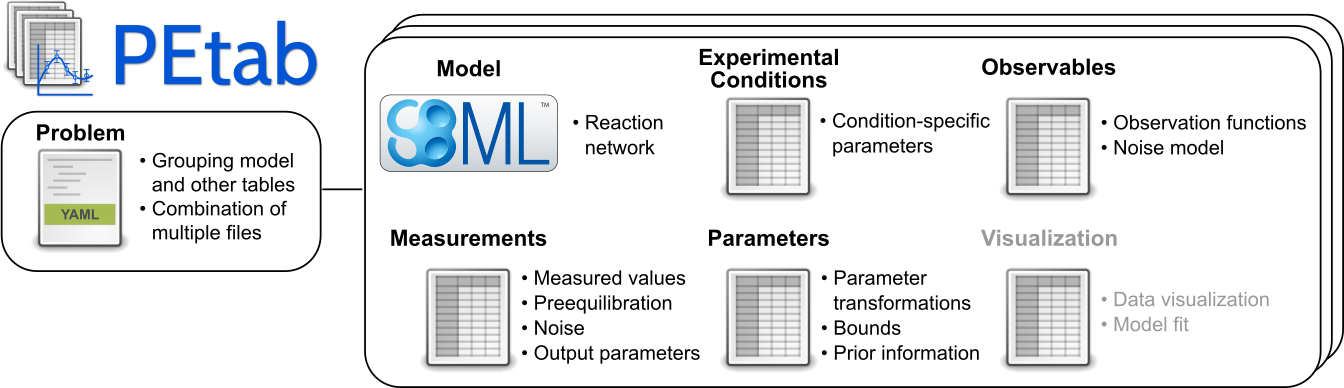
PEtab is built around SBML and based on tab-separated values (TSV) files. It is meant as a standardized way to provide information for parameter estimation, which is out of the current scope of SBML. This includes, for example:
Specifying and linking measurements to models
Defining model outputs
Specifying noise models
Specifying parameter bounds for optimization
Specifying multiple simulation conditions with potentially shared parameters
Documentation
Documentation of the PEtab data format is available at https://petab.readthedocs.io/en/latest/.
Contributing to PEtab
To participate in PEtab editor elections, discussions on new PEtab developments, or PEtab-related events, please join our petab-discuss mailing list.
Any contributions and feedback to PEtab are very welcome, see our contribution guide.
Examples
A wide range of PEtab examples can be found in the systems biology parameter estimation benchmark problem collection.
PEtab support in systems biology tools
For a list of tools supporting PEtab, see the software support page.
Using PEtab
If you would like to use PEtab yourself, please have a look at:
a PEtab tutorial going through the individual steps of setting up a parameter estimation problem in PEtab, independently of any specific software
the example models provided in the benchmark collection.
the tutorials provided with each of the softwares supporting PEtab
To convert your existing parameter estimation problem to the PEtab format, you will have to:
Specify your model in SBML.
Create a condition table.
Create a table of observables.
Create a table of measurements.
Create a parameter table.
Create a YAML file that lists the model and all of the tables above.
If you are using Python, some handy functions of the
PEtab library can help
you with that. This includes also a PEtab validator called petablint which
you can use to check if your files adhere to the PEtab standard. If you have
further questions regarding PEtab, feel free to post an
issue at our GitHub repository.
Editing PEtab files
PEtab files can be created or edited using any text editor or spreadsheet software, or programmatically (see libraries below). There is also a dedicated graphical user interface for creating and editing PEtab 1.0 files, called PEtab-GUI.
PEtab Python library
PEtab comes with a Python package for creating, checking, visualizing and working with PEtab files. This package is available at https://github.com/PEtab-dev/libpetab-python/.
PEtab R library
There is also an R package for PEtab in development, which currently supports: consistent manipulation of experimental conditions, measurements, and observables; and visualizing measurements. It also provides an interface to the PEtab Python library, and an interface to dMod that supports: conversion of dMod models to SBML; and simulation and visualization of results. The package is available at https://github.com/dlill/petab/.
PEtab extensions
Other standardization-related projects that extend or build on PEtab include:
PEtab Select PEtab Select brings model selection to PEtab. It comprises both a standardized way to define model selection problems in PEtab and a Python library to work with these problems.
petabunit (WIP) A Python library that extends PEtab to annotate and convert physical units.
PEtab SciML (WIP) PEtab SciML brings Scientific Machine Learning (SciML) models that combine mechanistic and machine learning models to PEtab. It comprises both a standardized way to define SciML problems, and a Python library to work with these problems.
Getting help
If you have any questions or problems with PEtab, feel free to post them at our GitHub issue tracker.
PEtab v1
PEtab v2
- PEtab v2 draft
- Purpose
- Scope
- Overview
- Changes from PEtab 1.0.0
- Model definition
- Condition table
- Experiment table
- Measurement table
- Observable table
- Parameter table
- Mapping table
- YAML file for grouping files
- Initialization and parameter application
- Frequentist vs. Bayesian inference
- Math expressions syntax
- Identifiers
- Extensions
- Tutorial